
Biomolecular condensates are membrane-less organelles that can selectively concentrate biomolecules, mainly proteins and nucleic acids. There are more than hundred unique biomolecular condensates known to date and their number is continuously rising (cd-code.org).
To aid the systematic discovery of condensate forming proteins, we created a machine learning based model called PICNIC (Proteins Involved in CoNdensates In Cells), that predicts proteins involved in biomolecular condensates regardless of the mechanism of condensate formation.
The first model (PICNIC) is based on sequence-based features and structure-based features derived from Alphafold2 models. Another model includes extended set of features based on Gene Ontology terms (PICNIC-GO). Although this model is biased by the already available annotations on proteins, it provides useful insights about specific protein properties that are enriched in proteins of biomolecular condensate. Overall, we recommend using PICNIC that is an unbiased predictor, and using PICNIC-GO for specific cases, for example for experimental hypothesis generation.
Although both models were trained on the human data, they generalize well to other organisms tested. We precomputed scores for the proteome of 14 species in Download.
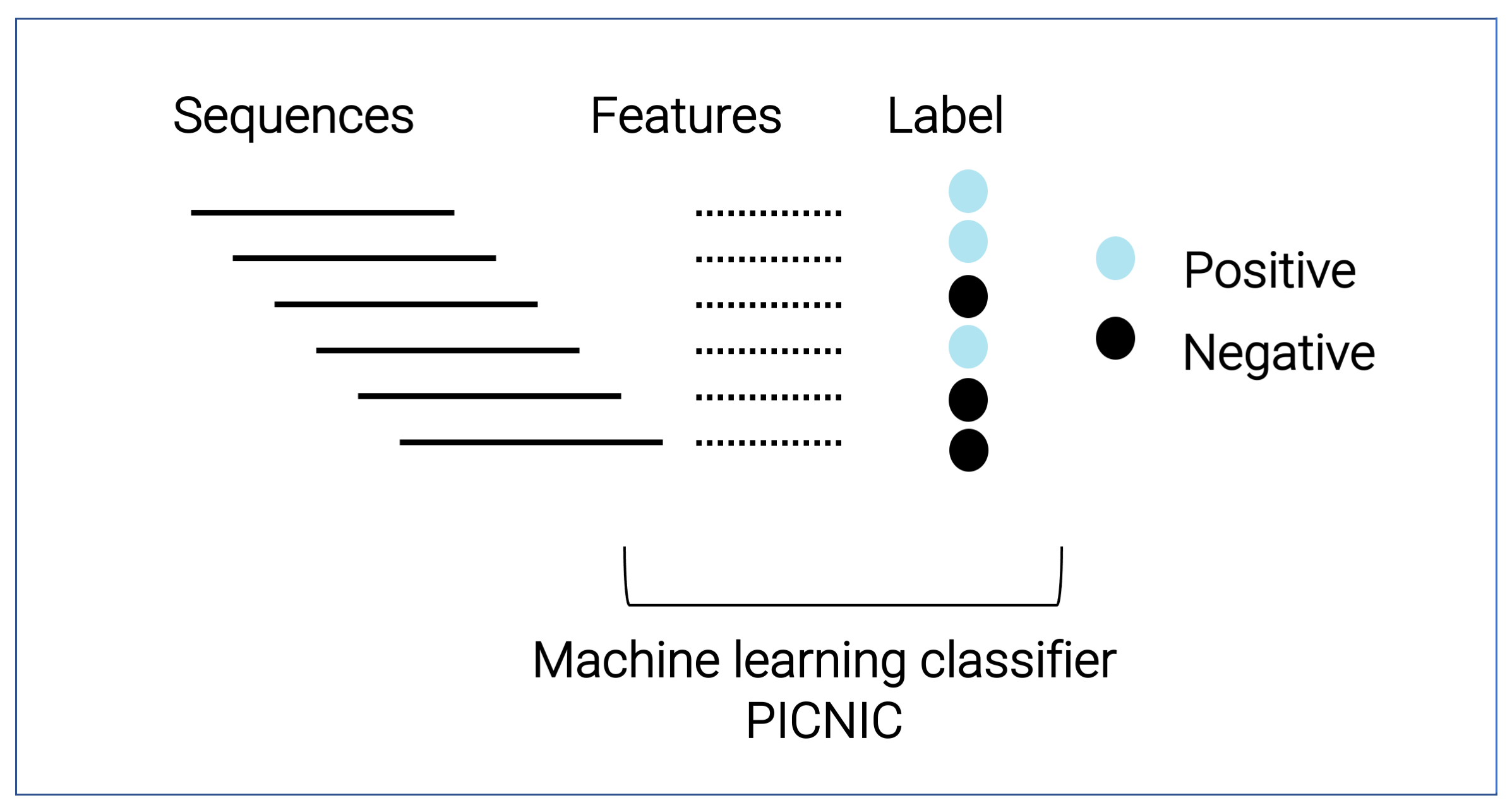
Pipeline of data driven machine learning algorithm
Gradient boosting machine was used as the model for machine learning classifier. It was trained on the set of features based on sequence, structure (predicted by Alphafold2) and function (reported by Gene Ontology annotation).
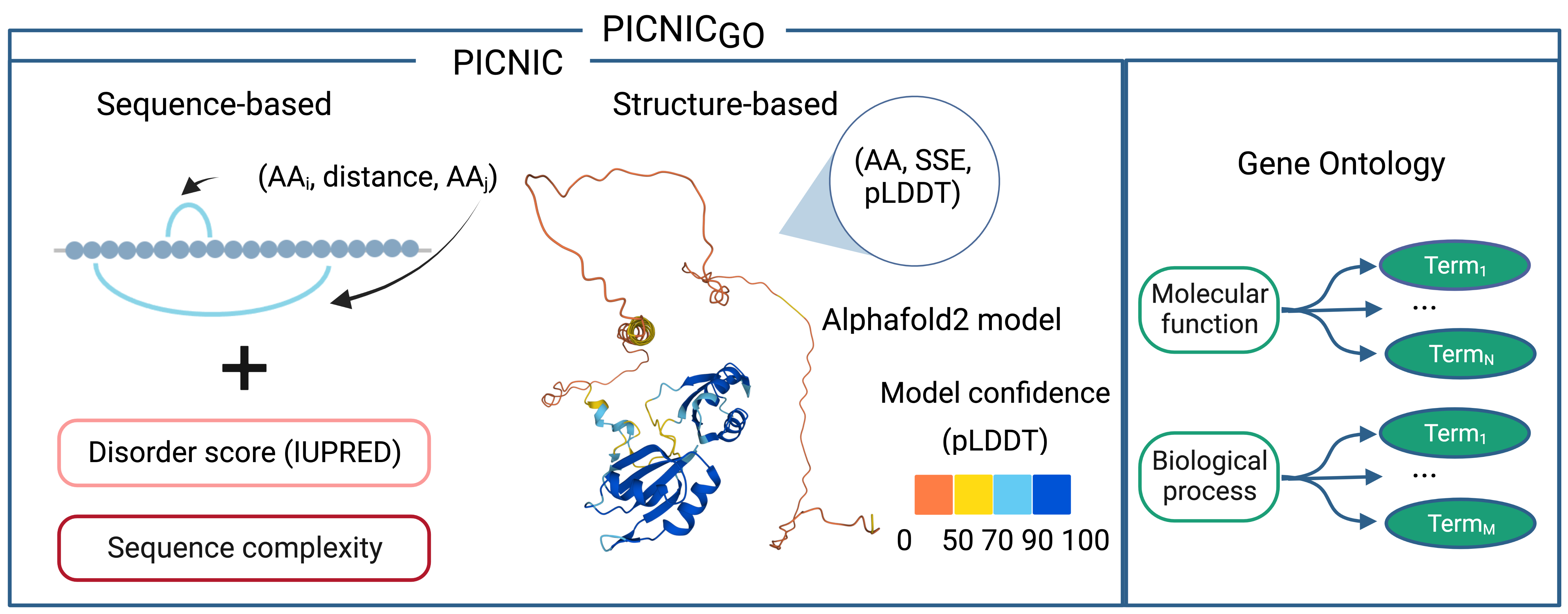
Features of machine learning classifier
Two types of models were developed: with (PICNIC-GO) and without the use of Gene Ontology annotation features (PICNIC). Sequence-based features include sequence complexity, disorder score (IUPRED), and features based on amino acid composition. The protein structures were downloaded from AlphaFold Protein Structure Database.
For more details on the model, see the publication: